Overview
Background
CEDARS (Clinical Event Detection and Recording System) is a computational paradigm for collection and aggregation of time-to-event data in retrospective clinical studies. Developed out of a practical need for a more efficient way to conduct medical research, it aims to systematize and accelerate the review of electronic health record (EHR) corpora to detect and characterize clinical events. Using CEDARS, human abstractors can work more efficently through the use of dedicated graphical user interface (GUI). the system also incorporates a customized data storage scheme and a natural language processing (NLP) pipeline. In its current iteration, CEDARS is available as an open-source Python package under GPL-3 license. The latest package and previous versions can be cloned from GitHub. Full documentation is available here. CEDARS can be used with or without its companion NLP package PINES. Please see the Terms of Use before using CEDARS. CEDARS is provided as-is with no guarantee whatsoever and users agree to be held responsible for compliance with their local government/institutional regulations.
General Requirements
- Python 3.9 or later
- Docker
Basic Concepts
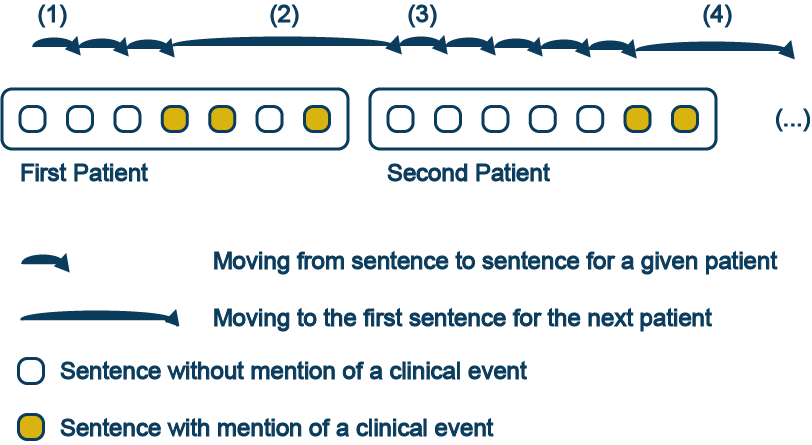
Sentences with keywords or concepts of interest are presented to the end user one at a time and in chronological order. The user assesses each sentence, determining whether or not a clinical event is being reported. The whole note or report drawn from the EHR is available for review in the GUI. If no event is declared in the sentence, CEDARS presents the next sentence for the same patient (#1). If an event date is entered, CEDARS moves to the next unreviewed sentence before the event date. If there are no sentences left to review before the event, the GUI moves to the next patient (#2) and the process is repeated with the following record (#3 and #4), until all selected sentences have been reviewed.
In order for CEDARS to be sufficiently sensitive and not miss and unacceptable number of clinical events, the keyword/concept search query must be well thought and exhaustive. The performance of CEDARS will vary by medical area, since the extent of medical lexicon will vary substantially between event types.
Features
Automatic Detection of Key EventsEasy interface to upload and query medical data.Project StatisticsSecure Login
Operational Schema
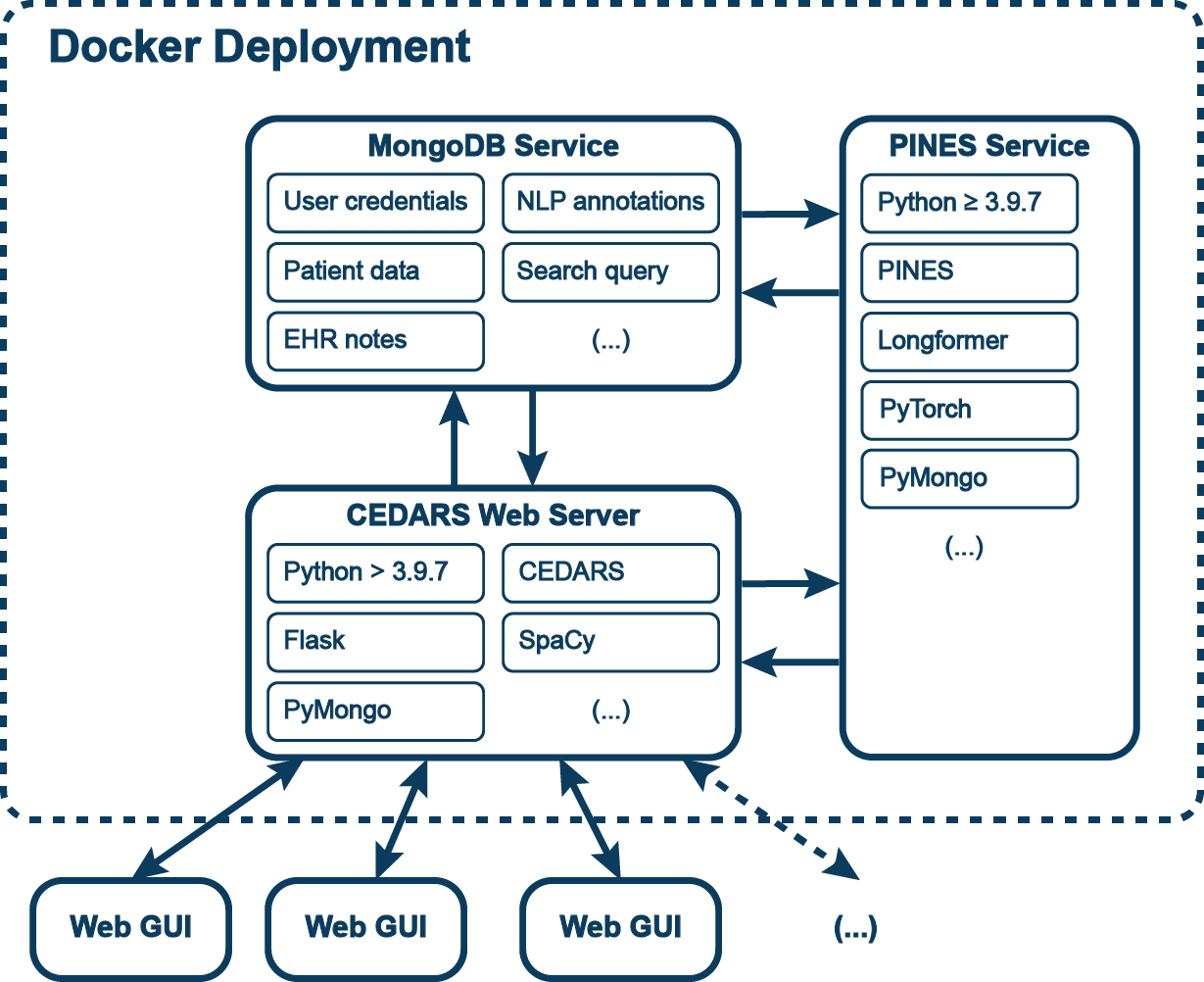
CEDARS is modular and all information for any given annotation project is stored in one MongoDB database. User credentials, original clinical notes, NLP annotations and patient-specific information are stored in dedicated collections. Once clinical notes have been uploaded, they are passed through the NLP pipeline. Currently only UDPipe is supported and integrated with CEDARS. If desired, the annotation pipeline can include negation and medical concept tagging by NegEx and UMLS respectively.
Multiple users can load the web GUI and annotate records at the same time. Once accessed, a given patient record is locked for the user.
Future Development
We are currently documenting the performance of CEDARS with a focus on oncology clinical research. At the present time, we wish to solidify the CEDARS user interface and ensure a smooth experience in multi-user settings. In the longer term, plug-in modules featuring enhanced query generation and adaptive learning will be integrated into the R workflow. Support for other NLP engines and extensive parallel processing are also desirable.
Please communicate with package author Simon Mantha, MD, MPH (smantha@cedars.io) if you want to discuss new features or using this software for your clinical research application.